Restructuring of bacteriophage ɸ6 viral particle activates semi-conservative transcription

Event details
| Date | 16.04.2024 |
| Hour | 12:15 › 13:15 |
| Speaker | Juha T. Huiskonen, Institute of Biotechnology, Helsinki Institute of Life Science HiLIFE, University of Helsinki, Finland |
| Location | |
| Category | Conferences - Seminars |
| Event Language | English |
Double-stranded RNA viruses transcribe their genome within a proteinaceous viral capsid to avoid host cell defenses. Members of the Reovirales use conservative transcription analogously to cellular gene expression. Most other dsRNA viruses, however, transcribe their genome using semi-conservative transcription whereby the positive strand of the genomic template becomes mRNA. Here, we visualize the activation of semi-conservative transcription in ɸ6 cystovirus particles using cryo-EM and Localized Reconstruction method (Ilca et al. 2015). The tri-segmented dsRNA genome is organized as a spool (Ilca et al. 2019). Upon transcription initiation, we observe a stepwise assembly of transcription complexes at the opposing poles of the genome spool, in addition to a concomitant expansion of the inner capsid layer and uncoating of the outer capsid layer. We show that the minor protein P7 is essential for transcription. The mature transcription complexes consist of asymmetric dimers of P7 dimers forming a triskelion around the viral polymerases (Fig. 1). The polymerases neighbor the vertices of the icosahedral capsid. The packaging hexamers at the vertices channel the viral mRNA exit. Our results define the complex molecular pathway from the quiescent state to the activation of semi-conservative intra-capsid transcription.
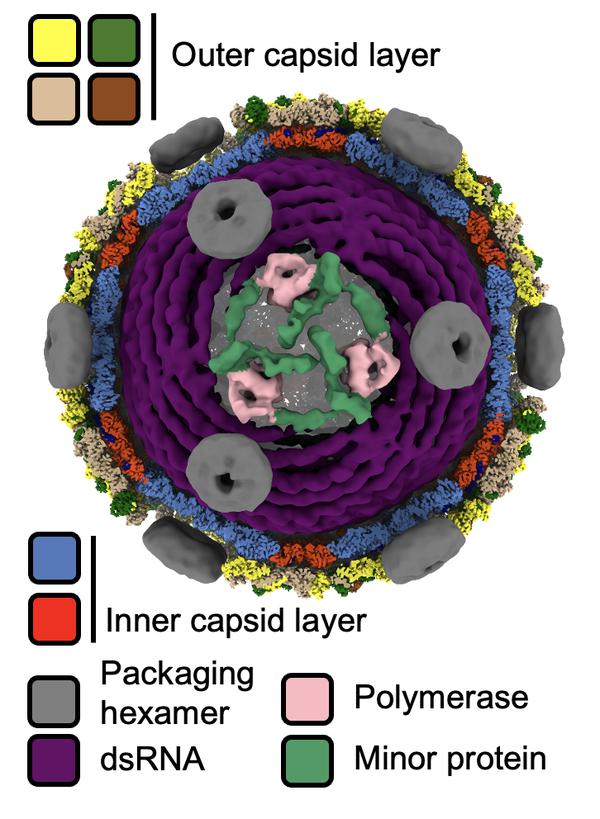
Fig. 1 dsRNA bacteriophage ɸ6 in transcribing state.
The viral particle is shown along the 3-fold axis of symmetry. The front half of the two capsid layers is removed to reveal the transcription initiation complex and the spooled dsRNA genome.
Serban L. Ilca1,2, Xiaoyu Sun3, Esa-Pekka Kumpula4, Katri Eskelin3, David I. Stuart2, Minna M. Poranen3, Juha T. Huiskonen2,4
1 New York Structural Biology Center, Simons Electron Microscopy Center, New York, NY, USA
2 Division of Structural Biology, Centre for Human Genetics, University of Oxford, Oxford, UK
3 Molecular and Integrative Biosciences Research Programme, Faculty of Biological and Environmental Sciences, University of Helsinki, Helsinki, Finland
4 Institute of Biotechnology, Helsinki Institute of Life Science HiLIFE, University of Helsinki, Helsinki, Finland
References:
Ilca, S.L., Kotecha, A., Sun, X., Poranen, M.M., Stuart, D.I., and Huiskonen, J.T. (2015). Localized reconstruction of subunits from electron cryomicroscopy images of macromolecular complexes. Nat Commun 6, 8843.
Ilca, S.L., Sun, X., El Omari, K., Kotecha, A., de Haas, F., DiMaio, F., Grimes, J.M., Stuart, D.I., Poranen, M.M., and Huiskonen, J.T. (2019). Multiple liquid crystalline geometries of highly compacted nucleic acid in a dsRNA virus. Nature 570, 252-256.
Keywords: semi-conservative transcription, cryo-EM, localized reconstruction, symmetry relaxation, dsRNA virus, bacteriophage
Supported by: Research Council of Finland and Sigrid Jusélius Foundation
Practical information
- Informed public
- Free
Organizer
- Aleksandar Antanasijevic